Goodness of Fit test between a distance based measure of spatial structure and simulations of its null hypothesis
GoFtest.RdCalculates the risk to reject the null hypothesis erroneously, based on the distribution of the simulations.
Usage
GoFtest(Envelope,
Test = "DCLF",
Scaling = "Asymmetric",
Range = NULL,
Alpha = 0.05,
CheckArguments = TRUE)Arguments
- Envelope
An envelope object (
envelope) containing simulations in itssimfunsattribute. It may be the result of any estimation function of the dbmss package or obtained by theenvelopefunction with argumentsavefuns=TRUE.- Test
A string specifying the method to summarize the deviation from the null hypothesis. The deviation may be summarized different ways:
"DCLF": the integrated squared deviation is utilized, a Diggle-Cressie-Loosmore-Ford test is performed (default);
"Integral": the integrated absolute deviation is utilized;
"MAD": the Maximum Absolute Deviation is utilized, a MAD test is performed.
- Scaling
A string specifying the method to scale the residuals of the test. Scaling may be:
"Asymmetric": the differences between the 2.5% lower quantiles of simulations and the expected value is utilized to scale negative residuals, and the differences between the 2.5% upper quantiles and the expected value is utilized to scale positive residuals (default);
"Quantile": ranges between the 2.5% upper and 2.5% lower quantiles is utilized to scale residuals, disregarding whether residuals are negative or positive;
"Studentized": the standard deviation of simulations is utilized.
"None": does not scale residuals.
- Range
A vector of length two containing the minimum and the maximum distance over which to compute the test. If
NULL, or if the selected range is outside the simulated distances, all distances in theEnvelopeargument are used.- Alpha
The risk level, 5% by default.
- CheckArguments
Logical; if
TRUE, the function arguments are verified. Should be set toFALSEto save time in simulations for example, when the arguments have been checked elsewhere.
Details
This function gathers multiple Goodness of Fit tests: the DCLF test (Diggle 1986, Cressie 1993, Loosmore & Ford 2006, Marcon et al. 2012, Myllymäki et al. 2015), the integrated deviation test (Diggle 1979), and the MAD test (Diggle 1979, Myllymäki et al. 2015).
Monte Carlo simulations assess how well observed distance-based measures of spatial structure align with expected measures under the null hypothesis, estimated here by the mean value of simulations. For both observed and simulated measures, residuals — calculated as the differences between observed and expected values — are computed at each distance r. These residuals are then transformed into test-specific statistics u:
Test = "MAD"uses the maximum absolute residual;Test = "Integral"andTest = "DCLF"use residuals to approximate the integrated deviation, and the integrated squared deviation.
A rank test on u evaluates the null hypothesis that the observed point pattern originates from the same point process as the simulations.
The unequal variance of the residuals at different intervals of r influences u statistics, and the power of Goodness of Fit tests.
Myllymäki et al. (2015) proposed to scale residuals using pointwise quantiles (Scaling = "Asymmetric", and Scaling = "Quantile"), and pointwise standard deviations (Scaling = "Studentized").
Goodness of Fit tests are sensitive to the distance interval over which they are performed. It is recommended to choose the distance interval to test based on a priori hypotheses (Wiegand & Moloney 2013, Baddeley et al. 2015).
References
Baddeley, A., Rubak, E., & Turner, R. (2015). Spatial Point Patterns: Methodology and Applications with R (0 ed.). Chapman and Hall/CRC. https://doi.org/10.1201/b19708
Cressie, N. A. C. (1993). Statistics for Spatial Data (1st ed.). Wiley. https://doi.org/10.1002/9781119115151
Diggle, P. J. (1979). On Parameter Estimation and Goodness-of-Fit Testing for Spatial Point Patterns. Biometrics, 35(1), 87. https://doi.org/10.2307/2529938
Diggle, P. J. (1986). Displaced amacrine cells in the retina of a rabbit: Analysis of a bivariate spatial point pattern. Journal of Neuroscience Methods, 18(1–2), 115–125. https://doi.org/10.1016/0165-0270(86)90115-9
Loosmore, N. B., & Ford, E. D. (2006). Statistical inference using the G or K point pattern spatial statistics. Ecology, 87(8), 1925–1931. https://doi.org/10.1890/0012-9658(2006)87[1925:SIUTGO]2.0.CO;2
Marcon, E., Puech, F., & Traissac, S. (2012). Characterizing the Relative Spatial Structure of Point Patterns. International Journal of Ecology, 2012, 1–11. https://doi.org/10.1155/2012/619281
Myllymäki, M., & Mrkvička, T. (2024). GET: Global Envelopes in R. Journal of Statistical Software, 111(3), 1-40. https://doi.org/10.18637/jss.v111.i03
Myllymäki, M., Grabarnik, P., Seijo, H., & Stoyan, D. (2015). Deviation test construction and power comparison for marked spatial point patterns. Spatial Statistics, 11, 19–34. https://doi.org/10.1016/j.spasta.2014.11.004
Wiegand, T., & Moloney, K. A. (2013). Handbook of Spatial Point-Pattern Analysis in Ecology. Chapman and Hall/CRC. https://doi.org/10.1201/b16195
Note
Ktest is a much better test (it does not rely on simulations) but it is limited to the K function against complete spatial randomness (CSR) in a rectangle window.
This test is inspired from Myllymäki et al. (2015), and a similar function deviation_test() exists in the R package GET (Myllymäki & Mrkvička, 2024)
Examples
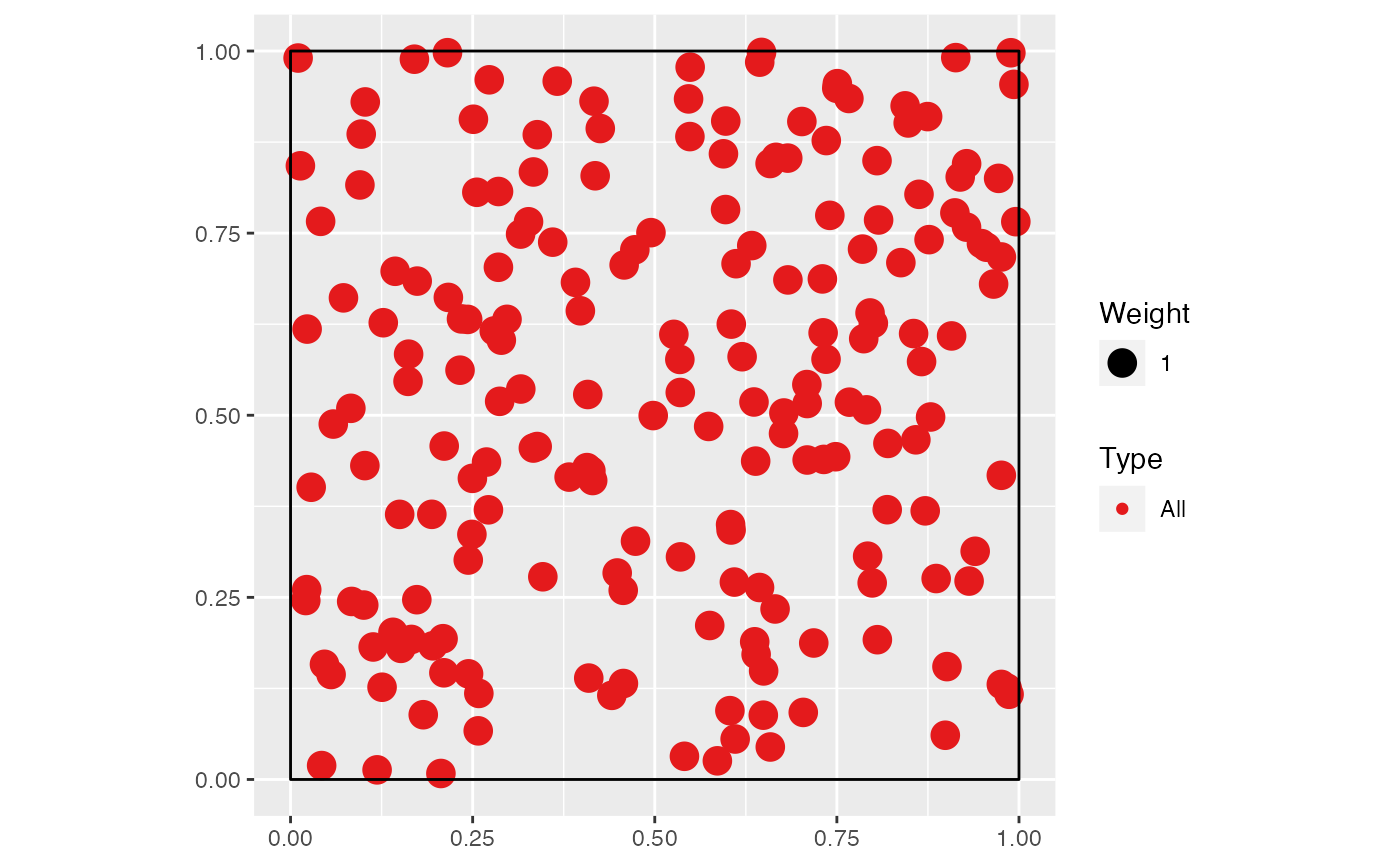
# Simulate a Matern (Neyman Scott) point pattern
nclust <- function(x0, y0, radius, n) {
return(runifdisc(n, radius, centre=c(x0, y0)))
}
X <- rNeymanScott(20, 0.2, nclust, radius=0.3, n=10)
autoplot(as.wmppp(X))
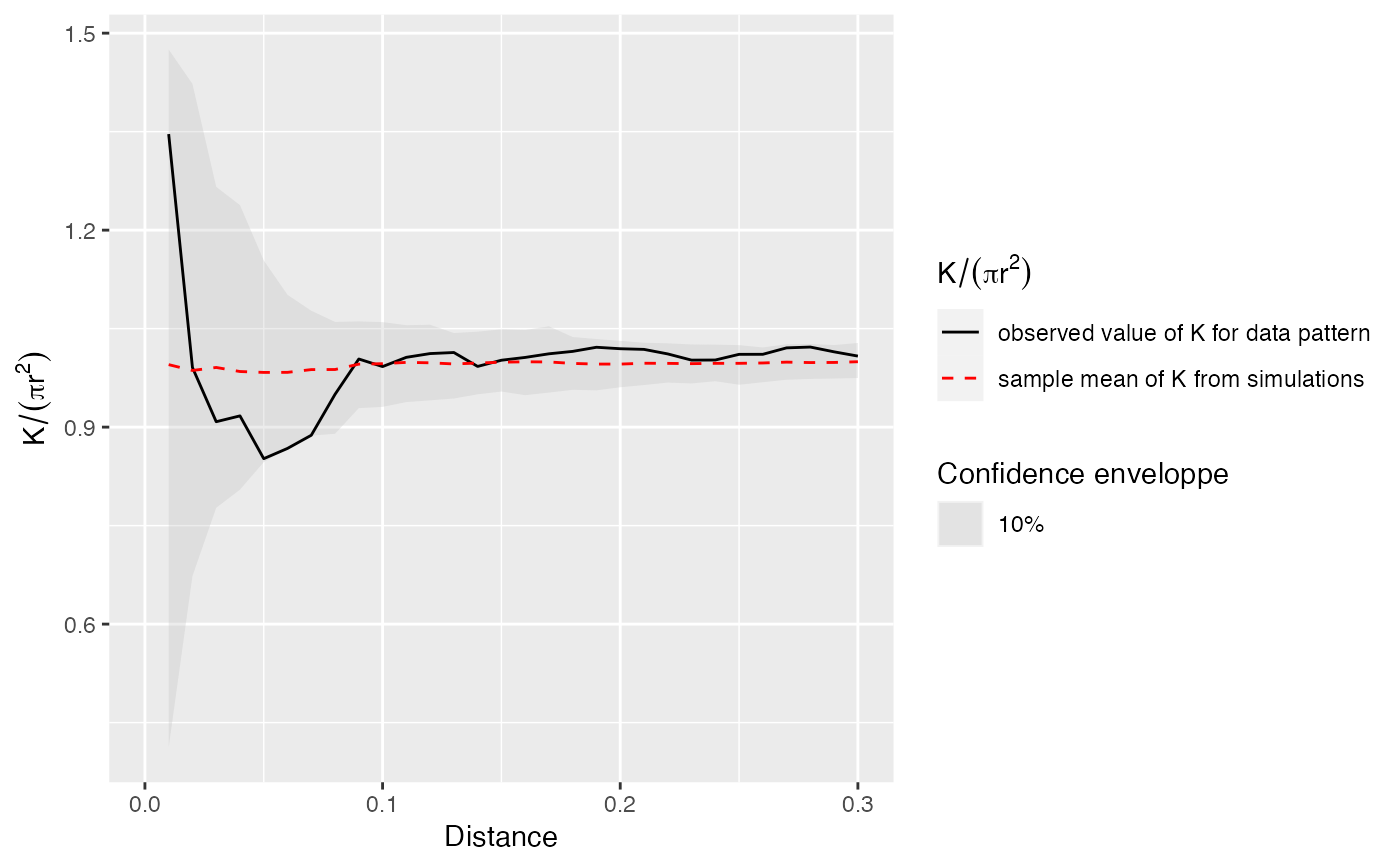
 # Calculate confidence envelope (should be 1000 simulations, reduced to 50 to save time)
r <- seq(0, 0.3, 0.01)
NumberOfSimulations <- 50
Alpha <- .10
Envelope <- KEnvelope(as.wmppp(X), r, NumberOfSimulations, Alpha)
autoplot(Envelope, ./(pi*r^2) ~ r)
# Calculate confidence envelope (should be 1000 simulations, reduced to 50 to save time)
r <- seq(0, 0.3, 0.01)
NumberOfSimulations <- 50
Alpha <- .10
Envelope <- KEnvelope(as.wmppp(X), r, NumberOfSimulations, Alpha)
autoplot(Envelope, ./(pi*r^2) ~ r)
 # DCLF test using asymmetric scaling.
# Power is correct if enough simulations are run (say >1000).
paste(
"p-value =",
GoFtest(Envelope, Test = "DCLF", Scaling = "Asymmetric", Range = c(0.1, 0.2))
)
#> Error in eval(expression(X), parent.frame()): object 'X' not found
# MAD test using asymmetric scaling.
paste(
"p-value =",
GoFtest(Envelope, Test = "MAD", Scaling = "Asymmetric", Range = c(0.1, 0.2))
)
#> Error in eval(expression(X), parent.frame()): object 'X' not found
# DCLF test using asymmetric scaling.
# Power is correct if enough simulations are run (say >1000).
paste(
"p-value =",
GoFtest(Envelope, Test = "DCLF", Scaling = "Asymmetric", Range = c(0.1, 0.2))
)
#> Error in eval(expression(X), parent.frame()): object 'X' not found
# MAD test using asymmetric scaling.
paste(
"p-value =",
GoFtest(Envelope, Test = "MAD", Scaling = "Asymmetric", Range = c(0.1, 0.2))
)
#> Error in eval(expression(X), parent.frame()): object 'X' not found