rcommunity() draws random communities according to a probability distribution.
rspcommunity() extends it by spatializing the random communities.
Usage
rcommunity(
n,
size = sum(abd),
prob = NULL,
abd = NULL,
bootstrap = c("Chao2015", "Marcon2012", "Chao2013"),
species_number = 300,
distribution = c("lnorm", "lseries", "geom", "bstick"),
sd_lnorm = 1,
prob_geom = 0.1,
fisher_alpha = 40,
coverage_estimator = c("ZhangHuang", "Chao", "Turing", "Good"),
check_arguments = TRUE
)
rspcommunity(
n,
size = sum(abd),
prob = NULL,
abd = NULL,
bootstrap = c("Chao2015", "Marcon2012", "Chao2013"),
species_number = 300,
distribution = c("lnorm", "lseries", "geom", "bstick"),
sd_lnorm = 1,
prob_geom = 0.1,
fisher_alpha = 40,
coverage_estimator = c("ZhangHuang", "Chao", "Turing", "Good"),
spatial = c("Binomial", "Thomas"),
thomas_scale = 0.2,
thomas_mu = 10,
win = spatstat.geom::owin(),
species_names = NULL,
weight_distribution = c("Uniform", "Weibull", "Exponential"),
w_min = 1,
w_max = 1,
w_mean = 15,
weibull_scale = 20,
weibull_shape = 2,
check_arguments = TRUE
)Arguments
- n
the number of communities to draw.
- size
the number of individuals to draw in each community.
- prob
a numeric vector containing probabilities.
- abd
a numeric vector containing abundances.
- bootstrap
the method used to obtain the probabilities to generate bootstrapped communities from observed abundances. If "Marcon2012", the probabilities are simply the abundances divided by the total number of individuals (Marcon et al. 2012) . If "Chao2013" or "Chao2015" (by default), a more sophisticated approach is used (see as_probabilities) following Chao et al. (2013) or Chao and Jost (2015) .
- species_number
the number of species.
- distribution
The distribution of species abundances. May be "lnorm" (log-normal), "lseries" (log-series), "geom" (geometric) or "bstick" (broken stick).
- sd_lnorm
the simulated log-normal distribution standard deviation. This is the standard deviation on the log scale.
- prob_geom
the proportion of resources taken by successive species of the geometric distribution.
- fisher_alpha
Fisher's \(\alpha\) in the log-series distribution.
- coverage_estimator
an estimator of sample coverage used by coverage.
- check_arguments
if
TRUE, the function arguments are verified. Should be set toFALSEto save time when the arguments have been checked elsewhere.- spatial
the spatial distribution of points. May be "Binomial" (a completely random point pattern except for its fixed number of points) or "Thomas" for a clustered point pattern with parameters
scaleandmu.- thomas_scale
in Thomas point patterns, the standard deviation of random displacement (along each coordinate axis) of a point from its cluster center.
- thomas_mu
in Thomas point patterns, the mean number of points per cluster. The intensity of the Poisson process of cluster centers is calculated as the number of points (
size) per area divided bythomas_mu.- win
the window containing the point pattern. It is an spatstat.geom::owin object. Default is a 1x1 square.
- species_names
a vector of characters or of factors containing the possible species names.
- weight_distribution
the distribution of point weights. By default, all weight are 1. May be "uniform" for a uniform distribution between
w_minandw_max, "weibull" with parametersw_min,weibull_scaleandshapeor "exponential" with parameterw_mean.- w_min
the minimum weight in a uniform, exponential or Weibull distribution.
- w_max
the maximum weight in a uniform distribution.
- w_mean
the mean weight in an exponential distribution (i.e. the negative of the inverse of the decay rate).
- weibull_scale
the scale parameter in a Weibull distribution.
- weibull_shape
the shape parameter in a Weibull distribution.
Value
rcommunity() returns an object of class abundances.
rspcommunity() returns either a spatialized community,
which is a dbmss::wmppp object , with PointType
values as species names if n=1 or an object of class ppplist
(see spatstat.geom::solist) if n>1.
Details
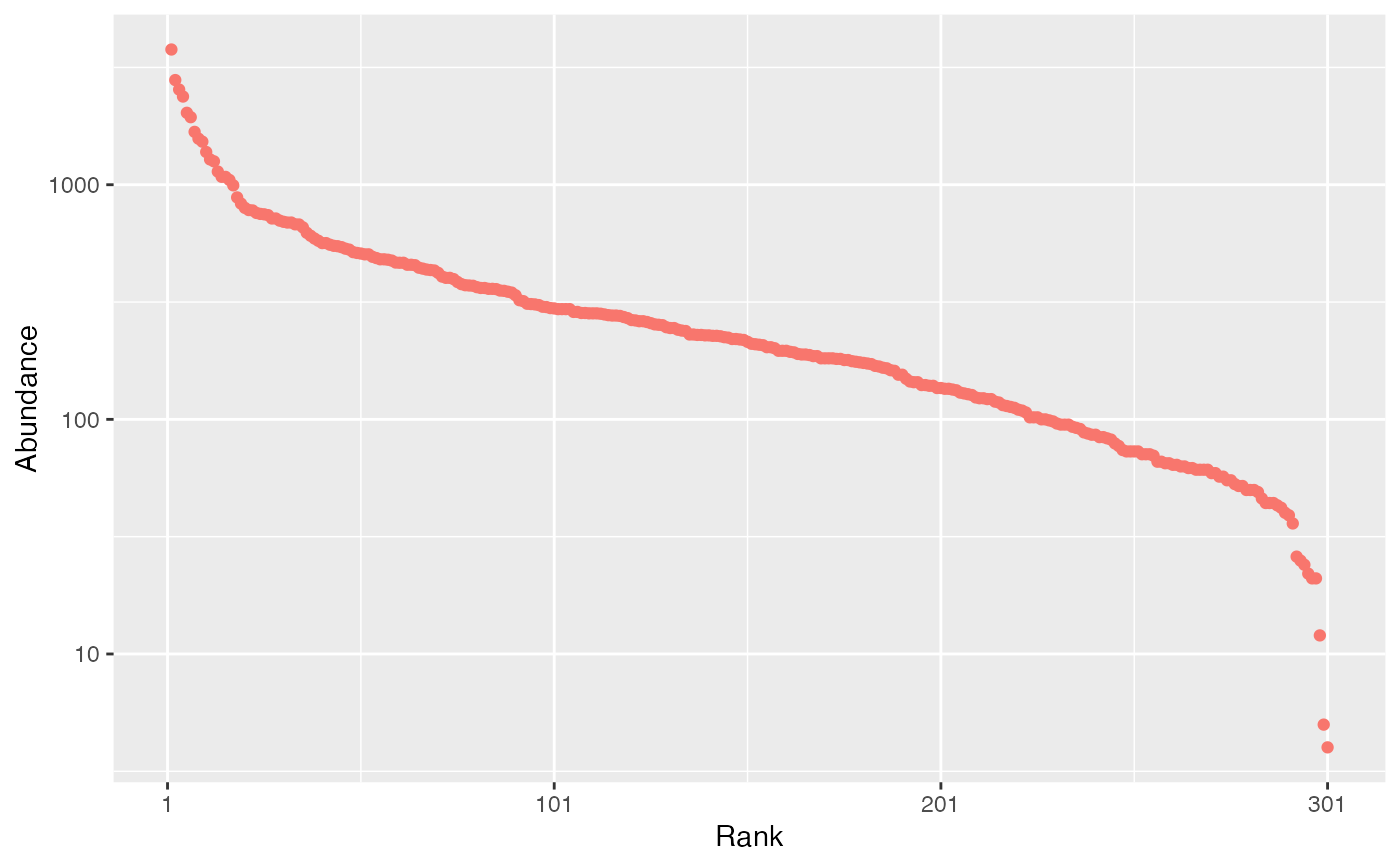
Communities of fixed size are drawn in a multinomial distribution according
to the distribution of probabilities provided by prob.
An abundance vector abd may be used instead of probabilities,
then size is by default the total number of individuals in the vector.
Random communities can be built by drawing in a multinomial law following
Marcon et al. (2012)
, or trying to estimate the
distribution of the actual community with probabilities.
If bootstrap is "Chao2013", the distribution is estimated by a single
parameter model and unobserved species are given equal probabilities.
If bootstrap is "Chao2015", a two-parameter model is used and unobserved
species follow a geometric distribution.
Alternatively, the probabilities may be drawn following a classical
distribution: either lognormal ("lnorm") (Preston 1948)
with given standard deviation (sd_lnorm; note that the mean is actually
a normalizing constant. Its value is set equal to 0 for the simulation of
the normal distribution of unnormalized log-abundances), log-series ("lseries")
(Fisher et al. 1943)
with parameter fisher_alpha, geometric
("geom") (Motomura 1932)
with parameter prob_geom,
or broken stick ("bstick") (MacArthur 1957)
.
The number of simulated species is fixed by species_number, except for
"lseries" where it is obtained from fisher_alpha and size:
\(S = \alpha \ln(1 + size / \alpha)\).
Note that the probabilities are drawn once only.
If the number of communities to draw, n, is greater than 1, then they are
drawn in a multinomial distribution following these probabilities.
Log-normal, log-series and broken-stick distributions are stochastic. The geometric distribution is completely determined by its parameters.
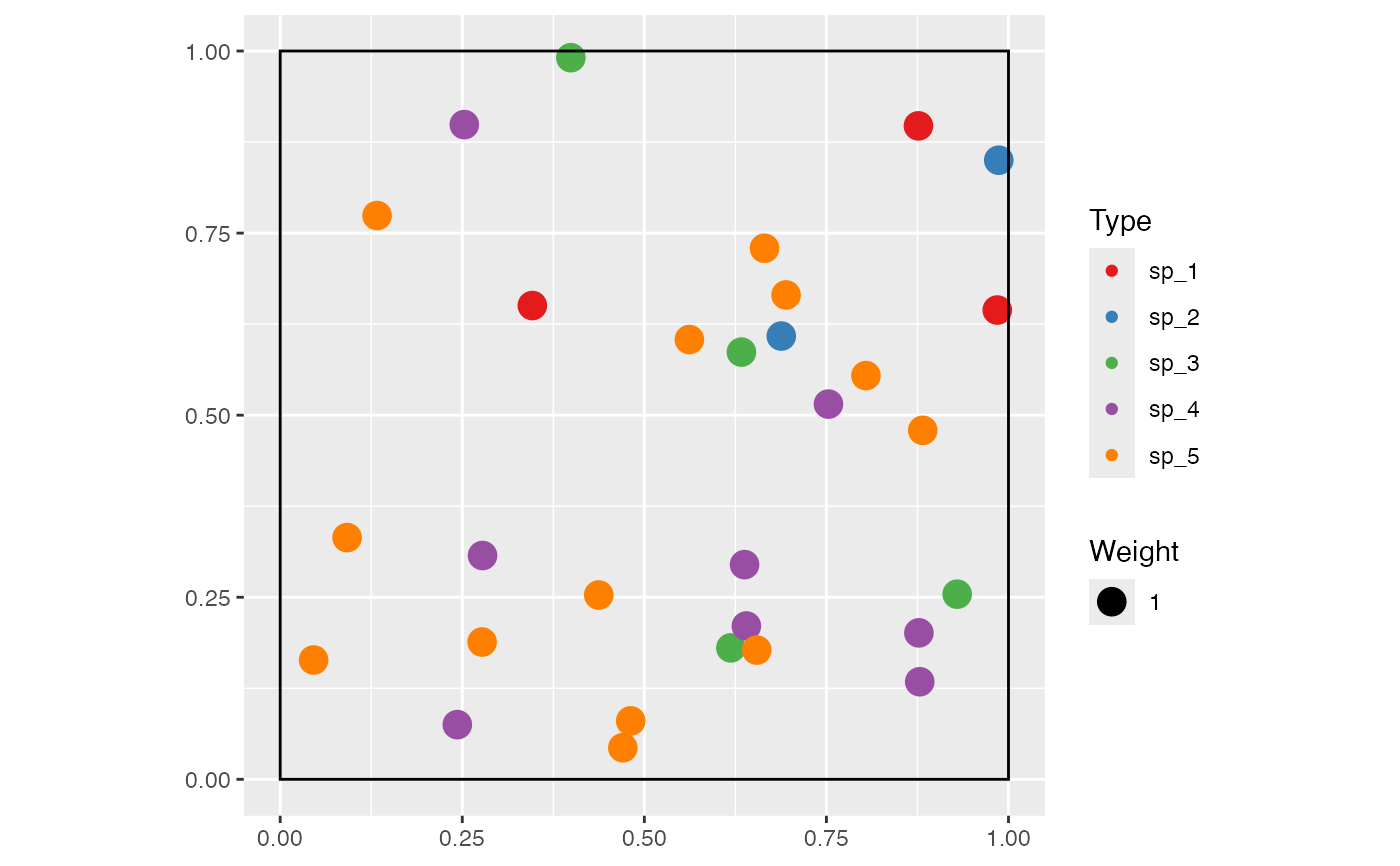
Spatialized communities include the location of individuals in a window, in a dbmss::wmppp object. Several point processes are available, namely binomial (points are uniformly distributed in the window) and Thomas (1949) , which is clustered.
Point weights, that may be for instance the size of the trees in a forest community, can be uniform, follow a Weibull or a negative exponential distribution. The latter describe well the diameter distribution of trees in a forest (Rennolls et al. 1985; Turner 2004) .
References
Chao A, Jost L (2015).
“Estimating Diversity and Entropy Profiles via Discovery Rates of New Species.”
Methods in Ecology and Evolution, 6(8), 873–882.
doi:10.1111/2041-210X.12349
.
Chao A, Wang Y, Jost L (2013).
“Entropy and the Species Accumulation Curve: A Novel Entropy Estimator via Discovery Rates of New Species.”
Methods in Ecology and Evolution, 4(11), 1091–1100.
doi:10.1111/2041-210x.12108
.
Fisher RA, Corbet AS, Williams CB (1943).
“The Relation between the Number of Species and the Number of Individuals in a Random Sample of an Animal Population.”
Journal of Animal Ecology, 12, 42–58.
doi:10.2307/1411
.
MacArthur RH (1957).
“On the Relative Abundance of Bird Species.”
Proceedings of the National Academy of Sciences of the United States of America, 43(3), 293–295.
doi:10.1073/pnas.43.3.293
, 89566.
Marcon E, Hérault B, Baraloto C, Lang G (2012).
“The Decomposition of Shannon's Entropy and a Confidence Interval for Beta Diversity.”
Oikos, 121(4), 516–522.
doi:10.1111/j.1600-0706.2011.19267.x
.
Motomura I (1932).
“On the statistical treatment of communities.”
Zoological Magazine, 44, 379–383.
Preston FW (1948).
“The Commonness, and Rarity, of Species.”
Ecology, 29(3), 254–283.
doi:10.2307/1930989
.
Rennolls K, Geary DN, Rollinson TJD (1985).
“Characterizing Diameter Distributions by the Use of the Weibull Distribution.”
Forestry, 58(1), 57–66.
ISSN 0015-752X, 1464-3626, doi:10.1093/forestry/58.1.57
.
Thomas M (1949).
“A Generalization of Poisson's Binomial Limit for Use in Ecology.”
Biometrika, 36(1/2), 18–25.
doi:10.2307/2332526
, 2332526.
Turner IM (2004).
The Ecology of Trees in the Tropical Rain Forest, 2nd edition.
Cambridge University Press.
ISBN 978-0-521-80183-6, doi:10.1017/CBO9780511542206
.