Transform a distance matrix into a similarity matrix (Leinster and Cobbold 2012) . Similarity between two species is defined either by a negative exponential function of their distance or by the complement to 1 of their normalized distance (such that the most distant species are 1 apart).
Arguments
- distances
a distance matrix or an object of class stats::dist.
- exponential
If
TRUE, similarity is \(e^{-r \delta}\), where \(r\) is argumentrate. IfFALSE, it is \(1 - \delta / \max(\delta)\).- rate
the decay rate of the exponential similarity.
- check_arguments
if
TRUE, the function arguments are verified. Should be set toFALSEto save time when the arguments have been checked elsewhere.
References
Leinster T, Cobbold C (2012). “Measuring Diversity: The Importance of Species Similarity.” Ecology, 93(3), 477–489. doi:10.1890/10-2402.1 .
Examples
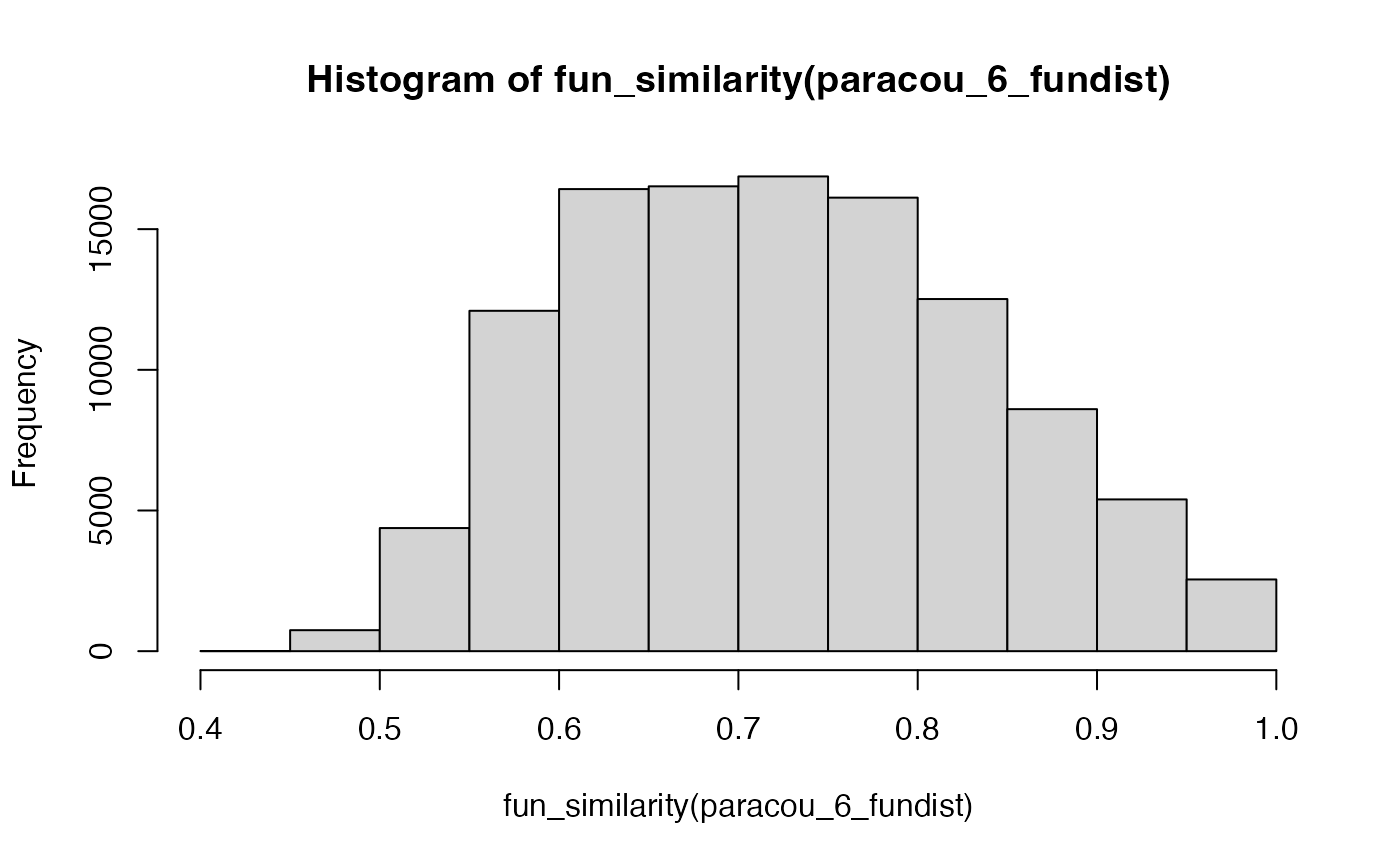
# Similarity between Paracou 6 species
hist(fun_similarity(paracou_6_fundist))