Diversity Partition of a metacommunity
DivPart.RdPartitions the diversity of a metacommunity into alpha and beta components.
Usage
DivPart(q = 1, MC, Biased = TRUE, Correction = "Best", Tree = NULL,
Normalize = TRUE, Z = NULL, CheckArguments = TRUE)
is.DivPart(x)
# S3 method for class 'DivPart'
plot(x, ...)
# S3 method for class 'DivPart'
autoplot(object, col = "grey35",
border = NA, ...)
# S3 method for class 'DivPart'
summary(object, ...)Arguments
- q
A number: the order of diversity. Default is 1.
- MC
A
MetaCommunityobject.- Biased
Logical; if
FALSE, a bias correction is appplied.- Correction
A string containing one of the possible corrections.
The correction must be accepted by
AlphaEntropy,BetaEntropyandGammaEntropy."Best"is the default value.- Tree
An object of class
hclust, "phylo" (seeread.tree),phylogorPPtree. The tree must be ultrametric.- Normalize
If
TRUE(default), diversity is not affected by the height of the tree.
IfFALSE, diversity is proportional to the height of the tree.- Z
A relatedness matrix, i.e. a square matrix whose terms are all positive, strictly positive on the diagonal. Generally, the matrix is a similarity matrix, i.e. the diagonal terms equal 1 and other terms are between 0 and 1.
- CheckArguments
Logical; if
TRUE, the function arguments are verified. Should be set toFALSEto save time when the arguments have been checked elsewhere.- x
An object to be tested or plotted.
- object
A
MCdiversityobject to be summarized or plotted.- col
The color used to fill the bars. See "Color Specification" in
par.- border
The color of the borders around the bars. See
rect.- ...
Additional arguments to be passed to the generic methods.
Details
DivPart partitions the diversity of the metacommunity into alpha and beta components. It supports estimation-bias correction.
If Tree is provided, the phylogenetic diversity is calculated else if Z is not NULL, then similarity-based entropy is calculated.
Beta diversity/entropy is calculated from Gamma and Alpha when bias correction is required, so community values are not available.
Value
A DivPart object. It is a list:
- MetaCommunity
The name of the
MetaCommunityobject containing inventory data.- Order
The value of
q.- Biased
Logical. If
FALSE, bias corrected values of diversity have been computed.- Correction
The estimation bias correction used to calculate diversity.
- Method
The method used to calculate entropy ("HCDT", "Similarity-based").
- Tree
The phylogenetic or functional tree used to calculate phylodiversity.
- Normalized
Logical. Indicates whether phylodiversity is normalized or proportional to the height of the tree.
- Z
The matrix used to calculate similarity-based entropy.
- TotalAlphaDiversity
The alpha diversity of communities.
- TotalBetaDiversity
The beta diversity of communities.
- GammaDiversity
The gamma diversity of the metacommunity.
- CommunityAlphaDiversities
A vector containing the alpha diversity of each community.
- TotalAlphaEntropy
The alpha entropy of communities.
- TotalBetaEntropy
The beta entropy of communities.
- GammaEntropy
The gamma entropy of the metacommunity.
- CommunityAlphaEntropies
A vector containing the alpha entropy of each community.
DivPart objects can be summarized and plotted.
References
Marcon, E., Herault, B., Baraloto, C. and Lang, G. (2012). The Decomposition of Shannon's Entropy and a Confidence Interval for Beta Diversity. Oikos 121(4): 516-522.
Marcon, E., Scotti, I., Herault, B., Rossi, V. and Lang, G. (2014). Generalization of the partitioning of Shannon diversity. PLOS One 9(3): e90289.
Marcon, E., Herault, B. (2015). Decomposing Phylodiversity. Methods in Ecology and Evolution 6(3): 333-339.
Marcon, E., Zhang, Z. and Herault, B. (2014). The decomposition of similarity-based diversity and its bias correction. HAL hal-00989454(version 3).
Examples
# Load Paracou data (number of trees per species in two 1-ha plot of a tropical forest)
data(Paracou618)
# Estimate Shannon diversity.
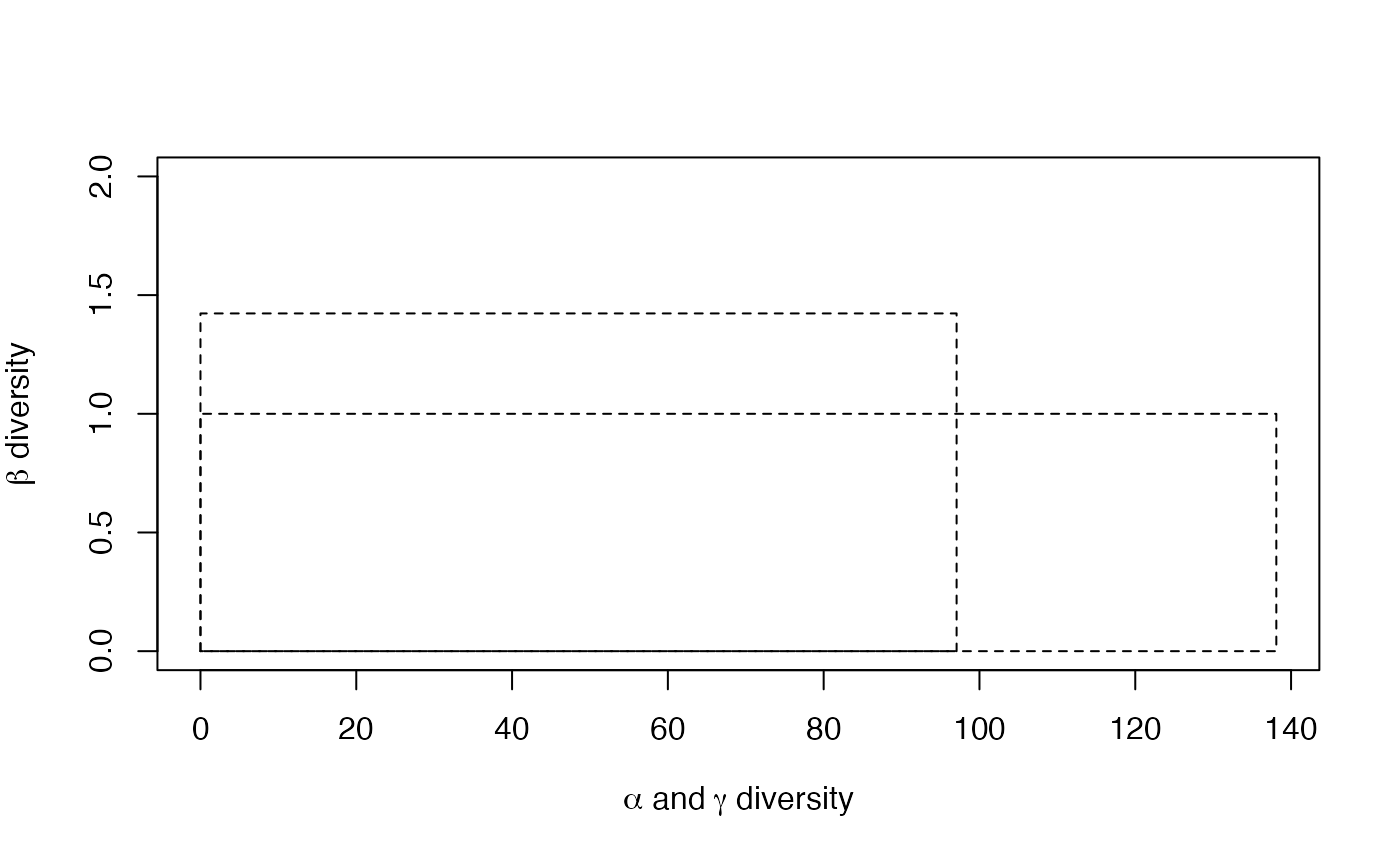
summary(DivPart(q = 1, Paracou618.MC, Biased = FALSE) -> dp)
#> HCDT diversity partitioning of order 1 of metaCommunity Paracou618.MC
#> with correction: UnveilJ
#> Alpha diversity of communities:
#> P006 P018
#> 83.7268 118.2713
#> Total alpha diversity of the communities:
#> [1] 97.06467
#> Beta diversity of the communities:
#> UnveilJ
#> 1.422843
#> Gamma diversity of the metacommunity:
#> UnveilJ
#> 138.1078
plot(dp)