Preprocessed Trees.
PPtree.RdMethods for objects of type "PPtree".
Usage
is.PPtree(x)
# S3 method for class 'PPtree'
plot(x, ...)Value
An object of class PPtree is a list:
- phyTree
A "phylo" (see
read.tree) tree- hTree
A
hclusttree- Height
The height of the tree, that is to say the distance between root and leaves
- Cuts
A vector. Cut times of the tree (the distance from nodes to leaves)
- Intervals
A vector. The lengths of intervals between cuts
is.PPtree returns TRUE if the object is of class PPtree.
plot.PPtree plots it.
Note
Versions up to 1.3 contained a phylog tree, now deprecated in ade4. A "phylo" (see read.tree) tree is now used.
See the dedicated vignette (vignette("Phylogenies", package="entropart")) for more details.
Examples
data(Paracou618)
# Preprocess a phylog object
ppt <- Preprocess.Tree(EightSpTree)
# Is it a preprocessed tree?
is.PPtree(ppt)
#> [1] TRUE
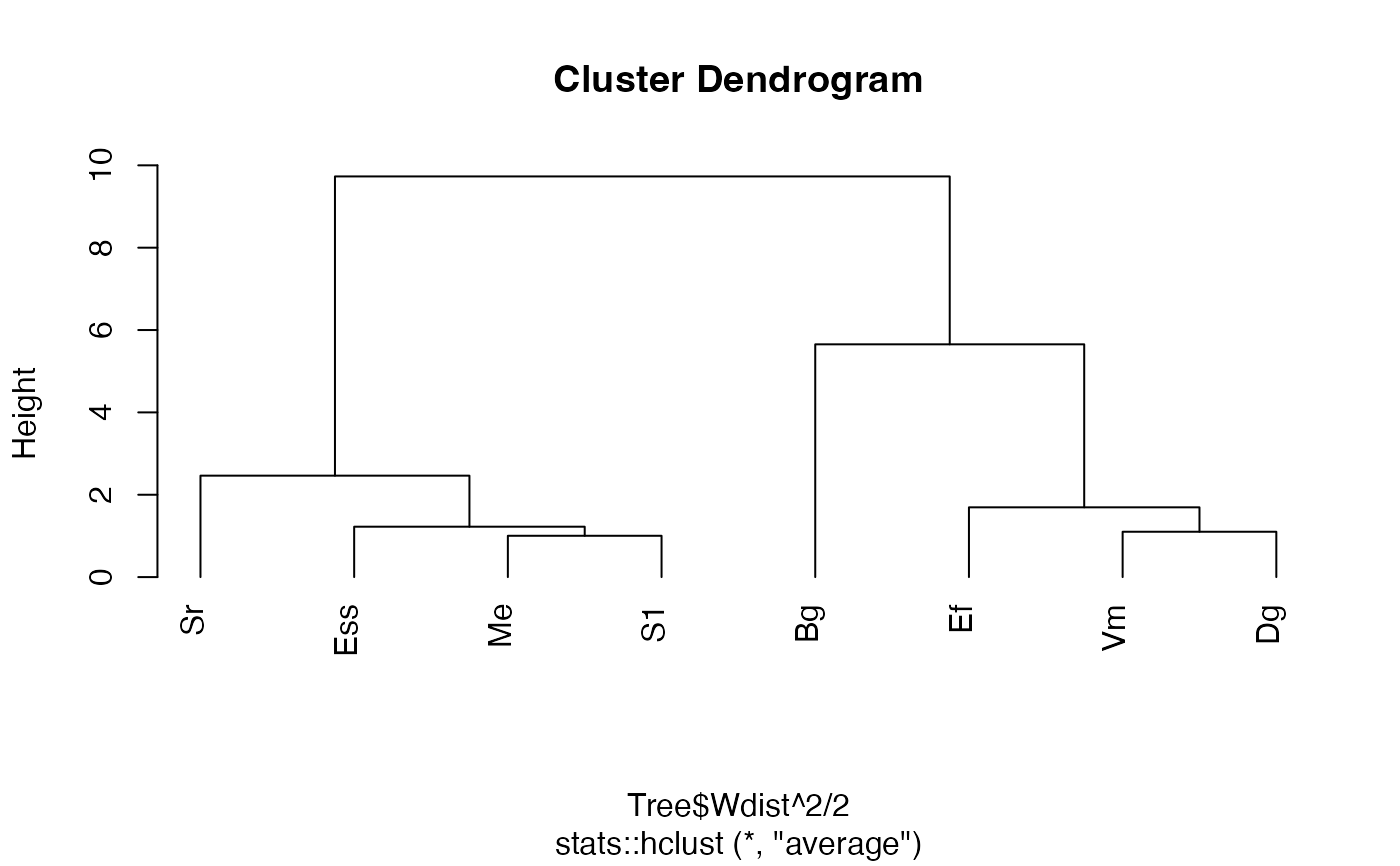
# Plot it
plot(ppt, hang=-1)
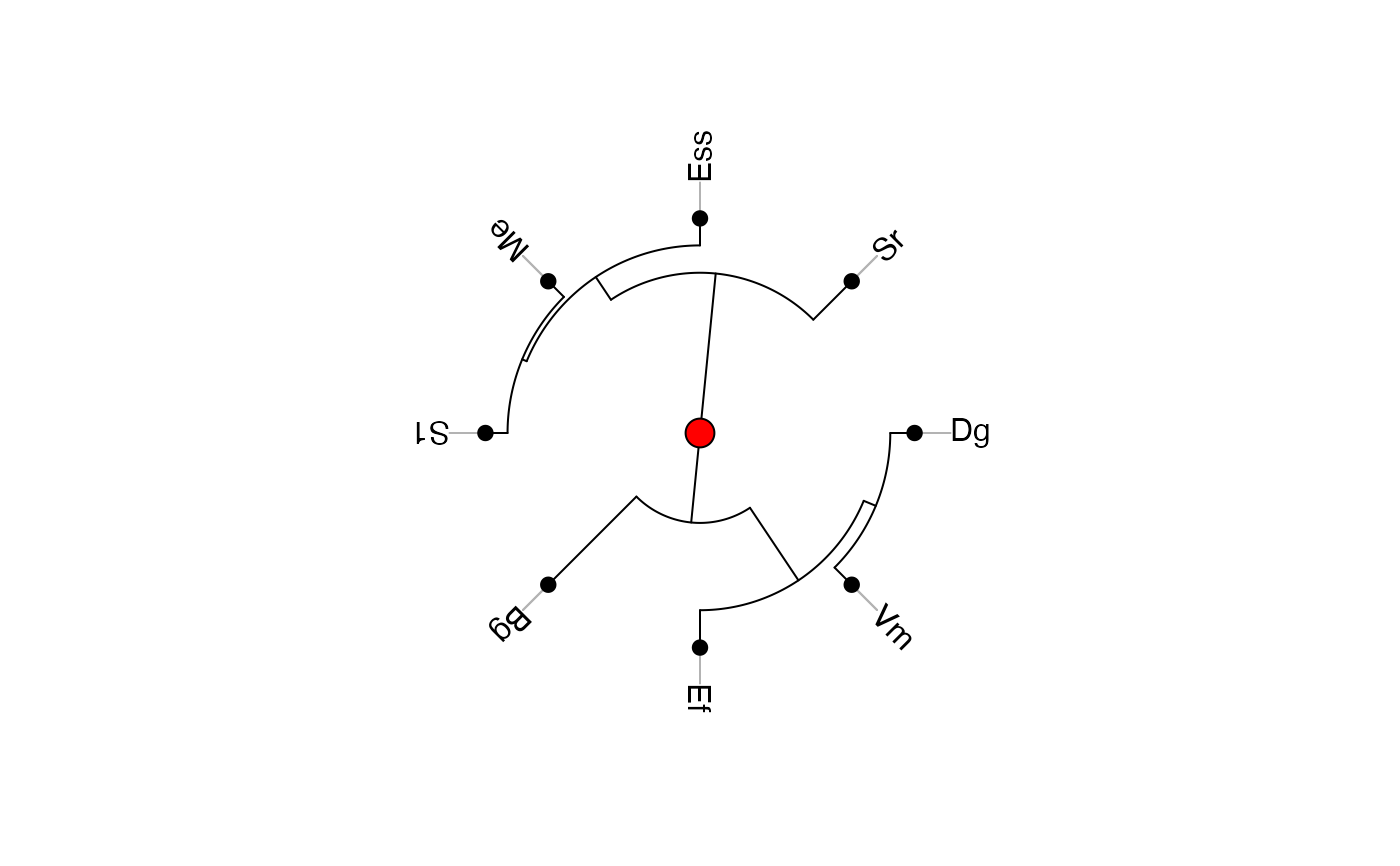
 # Alternative plot
ade4::radial.phylog(EightSpTree)
# Alternative plot
ade4::radial.phylog(EightSpTree)